We are excited to share this press release from No Stomach For Cancer Scientific Advisory Council member Raquel Seruca about their latest paper on germline missense mutations. According to Dr. Seruca, this paper describes a new tool of analyses to be part of a pipeline that helps with the decision of putative the clinical impact of this type of germline alterations.
Of great significance in this news is that, in addition to improving the care of HDGC patients, their findings can be applied to other cancers where e-cadherin mutations are known to be involved, including breast, colorectal, thyroid, and ovarian cancers.
The paper was published in the European Journal of Human Genetics.
Quantification of mutant E-cadherin using bioimaging analysis of in situ fluorescense microscopy. A new approach to CDH1 missense variations.
Dr. Seruca is one of many dedicated members of an international group who gathered at Radboud University Medical Centre in Nijmegen, The Netherlands in March 2014 to update the current consensus guidelines for HDGC that will be published in the coming months.

Raquel Seruca, MD, PHD
Group Leader of Cancer Genetics Group
Institute of Molecular Pathology and Immunology of Porto University (IPATIMUP)
Affiliated Professor, Medical Faculty of Porto University, Portugal
Press Release
2D images as the new tool for cancer prevention
30 November 2014 Ciência Viva – Agência Nacional para a Cultura Científica e Tecnológica
Portuguese researchers have developed a new method, which uses images of a protein in cells to quantify its distribution (how much protein there is, and where it is in the cell) for that population of cells. The discovery by researchers João Sanches, Raquel Seruca, and colleagues has important medical implications because the cellular location of a protein is directly linked to its function, as proved when the new algorithm identified mutations that had the potential to lead to hereditary diffuse gastric cancer (HDGC). This result is particularly important because HDGC is asymptomatic and, at the moment, its early detection is not reliable, which means that the disease has an extremely high mortality. The new algorithm with its ability to identify patients at high risk, hopefully, will change that.
The study, out in the European Journal of Human Genetics, is a collaboration between João Sanches from the Institute for Systems and Robotics and the Department of Bioengineering from the Instituto Superior Técnico at the Technical University of Lisbon that developed the algorithm, and the group of Raquel Seruca from the Department of Cancer Genetics of IPATIMUP – Institute of Molecular Pathology and Immunology at the University of Porto that works in cancer.
Gastric cancer is the 4th most common cause of cancer in the world, and hereditary diffuse gastric cancer (HDGC) makes up to 3% of all cases. Although not very frequent, the disease is hugely problematic for clinicians because of its high mortality, which is the result of several issues.
The first is the fact that HDGC is caused by functional abnormalities/mutations in e-cadherin, an adhesion protein of epithelial cells (those covering the surfaces and inside of the body). E-cadherin holds epithelial cells together by lying across the cell membrane, one end attached to e-cadherins from neighbouring cells and the other to the skeleton of the epithelial cell. When e-cadherin stops working, the cells become loose. In the case of HDGC, this means that there is no solid tumour, but, instead, a loose layer of cancerous cells, which easily move, spreading the disease and making HDGC control impossible very quickly. This is also why e-cadherin is considered an important tumour suppressor molecule.
The second problem lies in the difficulty of spotting HDGC early. Its initial symptoms are very non-specific (stomach acidity and burping for example), so they are easy to miss. And without clear symptoms, early detection relies solely on analysis of gastric cells looking for a loss of e-cadherin on the membrane (a malfunctioning e-cadherin also leaves the membrane to go inside the cell to be destroyed) trying to catch the early steps of the disease. But visual detection is not a reliable control method. To make things worse, not all e-cadherin mutations can cause HDGC, some are neutral/innocuous (meaning that they do not affect its function) making the identification and monitoring of those at risk of disease even more problematic.
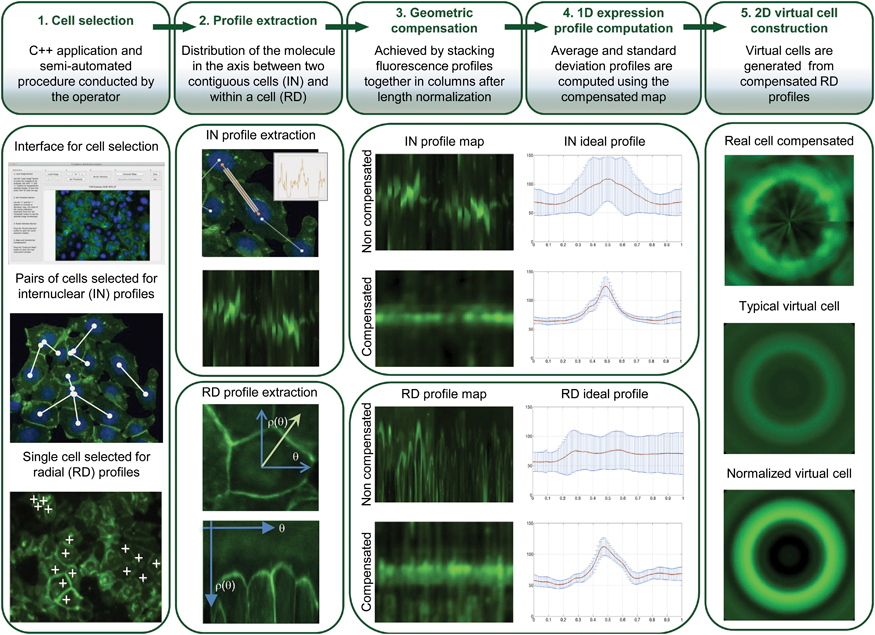
So there has been a major effort to improve HDGC detection, and it is here that Sanches’ algorithm steps in. Their algorithm brings many advantages – it is semi-automated which allows on one hand the human operators to resort to their experience to choose the more representative cells, and on the other to rely on the computer to normalize results from cells with very different sizes and shapes to work even with very heterogeneous cell populations. Resorting to images means that the cells are hardly disturbed. All these details ensure that the results are extremely accurate and representative of what is happening in live cells/individuals.
But how does the method actually work?
The software starts by collecting multiple fluorescence images of the protein in a cell population, and from their data generates “maps” of the protein distribution for that population. From these, it then constructs a 2D virtual image of a “typical cell” (for that specific population), and calculates a new parameter called maximum mean ratio (MMR), which quantifies the sharpness of the protein fluorescence peak (so effectively measures the protein quantity). In HDGC experiments, a high MMR meant that e-cadherin has high expression by the membrane (as it should), and low levels inside of the cell.
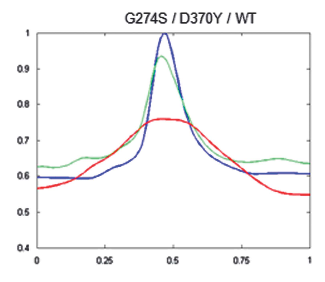
To see if this data could be used to improve the clinical management of HDGC, next the researchers used the algorithm to compare cells with working e-cadherins, with those carrying mutations that made the protein non-functional/useless (so cancer prone). The idea was, that since normal e-cadherin is on the membrane and its malfunctioning form moves inside the cell to be destroyed, the new algorithm, by providing the location of e-cadherin, should be able to identify individuals at risk of HDGC.
And in fact, after constructing 2D images of the two types of cells, the distinction was clear. Normal cells (or those with neutral/innocuous mutations) produced an image of a fluorescence circle with a clear centre (which represented the absence of protein inside the cell), while those with malfunctioning e-cadherin (that can lead to HDGC) showed a full fluorescence circle (the fluorescence in the centre represented e-cadherin inside cell). Additionally, these last cells had much lower MMR than normal, meaning less e-cadherin by the membrane and, in consequence, a weaker cell-to-cell adhesion which agreed with their propensity to cancer. “Our algorithm was not only able to pinpoint the protein location, but also to quantify it in each cellular compartment. While the MMR value gave us the protein dispersion.” – resumed Seruca
In conclusion, with a combination of 2D images and quantitative “maps” it should now be much easier for researchers (and in the future clinicians) to quickly and reliably identify those individuals with mutations that lead e-cadherin to lose their function, and who, as result, are prone to develop HDGC and should be put under close monitoring for early signs of disease.
“Our algorithm can now be used as a complementary approach to evaluate the pathogenicity of E-cadherin mutations.”- says Seruca – “Moreover, it can be applied to a wide range of proteins and, more importantly, to diseases characterized by aberrant protein expression or trafficking deregulation. “
And although the study has focused on HDGC, e-cadherin mutations are known to be involved in a variety of other cancers including breast, colorectal, thyroid, and ovarian so these new results could also be applied to them.